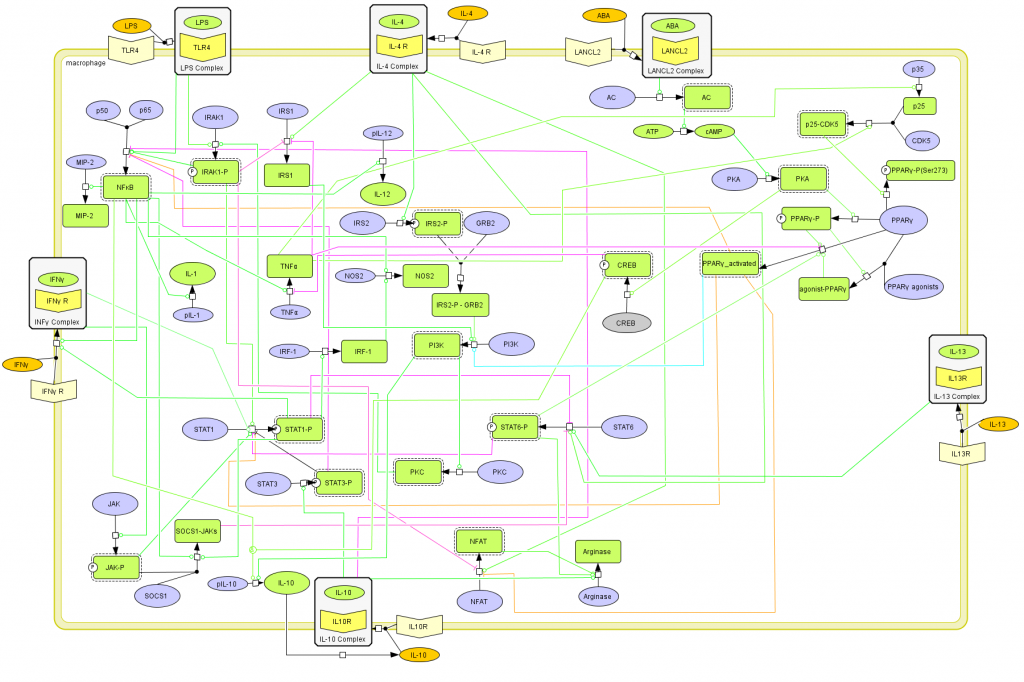
This is a version of MIEP Macrophage computational model released on April 28th, 2012. The model is available for download in CellDesigner xml format. We have tested that the model is compatible with Cell Designer 4.1.
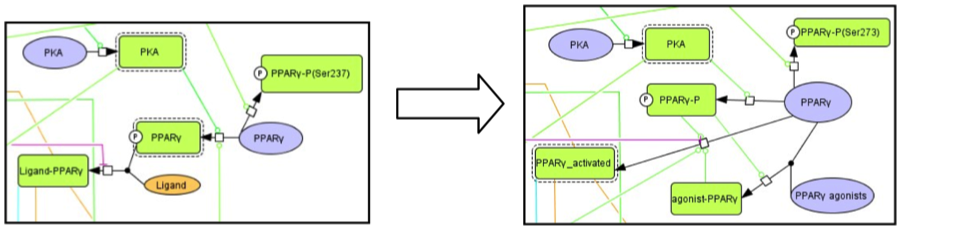
Changes were made focusing on PPAR gamma activation manner. In the modified model, PPAR gamma can be activated either by agonist binding or phosphorylation by PKA. Furthermore, there are two possible mechanisms of PPAR gamma activation by PKA phosphorylation: ligand-independent or –dependent manners.
The following is the structure figure of the model, and by clicking on the figure you can navigate the model through a Google-Map-API-enabled CellPublisher user friendly interface.
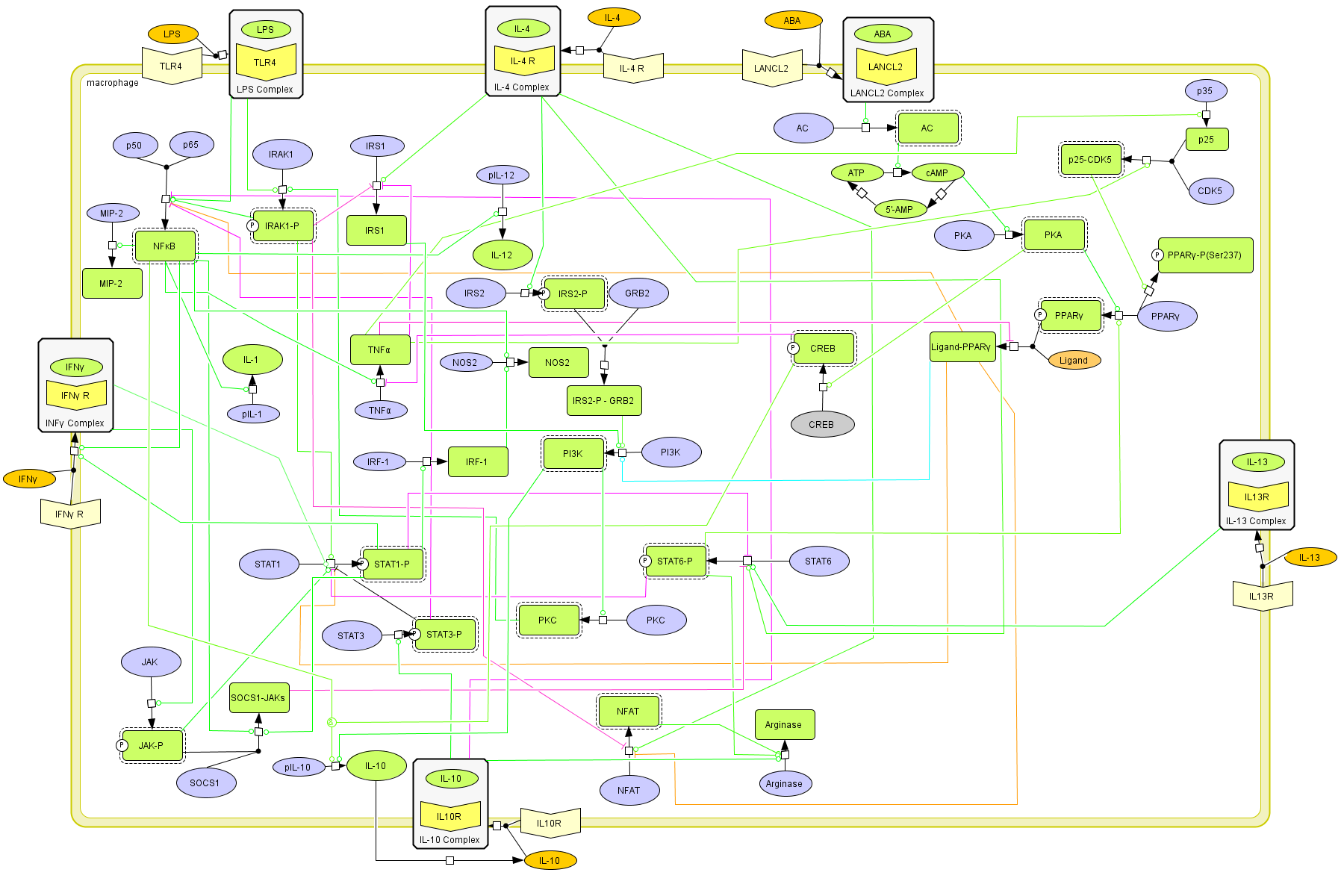
This is a version of MIEP Macrophage computational model released on Jan 11th, 2011. The model is available for download in CellDesigner xml format. We have tested that the model is compatible with Cell Designer 4.1. Here is the release note: One species and one reaction were added in the model. The new specie is s140. The reaction is re89. The aim to add these two items is to distinguish different phosphorylations of PPAR gamma. PKA phosphrylates PPAR gamma that increases PPAR gamma activity, while Cdk5 phosphrylates PPAR gamma that decrease PPAR gamma activity.
The following is the structure figure of the model, and by clicking on the figure you can navigate the model through a Google-Map-API-enabled CellPublisher user friendly interface.
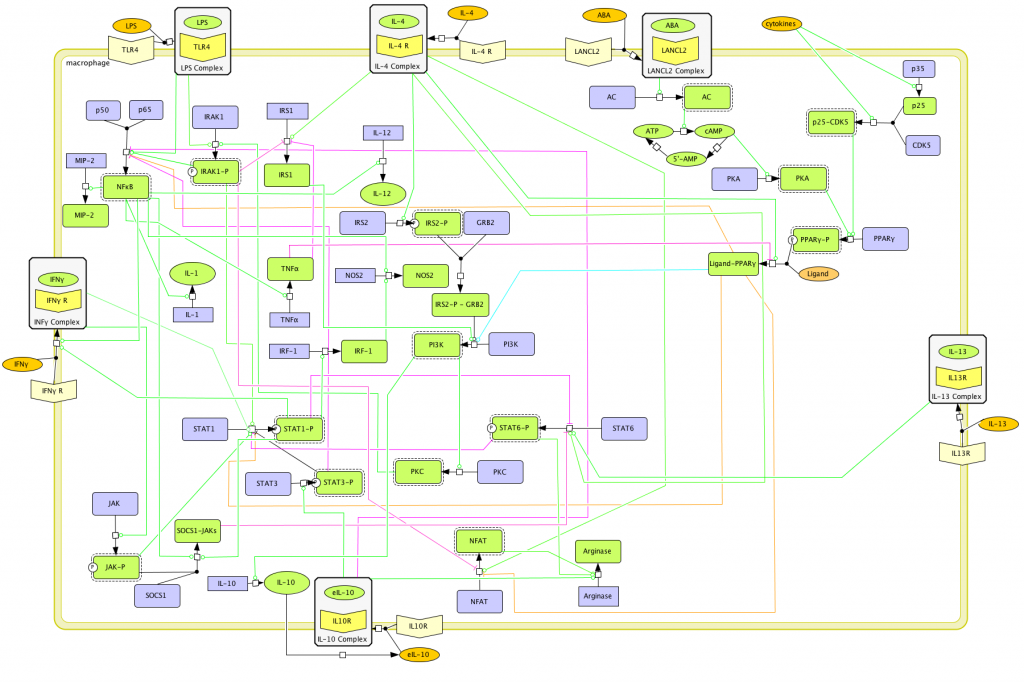
This is a version of MIEP Macrophage computational model released on Oct 7th, 2011. The model is available for download in CellDesigner xml format. We have tested that the model is compatible with Cell Designer 4.1. The following is the structure figure of the model, and by clicking on the figure you can navigate the model through a Google-Map-API-enabled CellPublisher user friendly interface.