Bistability analyses of immune responses to Helicobacter pylori infection
Helicobacter pylori is a dominant member of the stomach microbiome in large portions of the world’s population. Its presence has been associated with beneficial health effects in allergic and metabolic diseases, specifically asthma and Type 2 Diabetes. However, an estimated 15% of individuals colonized by H. pylori will develop gastric inflammation leading to abdominal pain, ulcers, or gastric cancer. As a result of these severe side effects, standard treatment involves eradication of the bacterium with antimicrobials, which have been linked to resistance and destruction of beneficial gut microbiota.
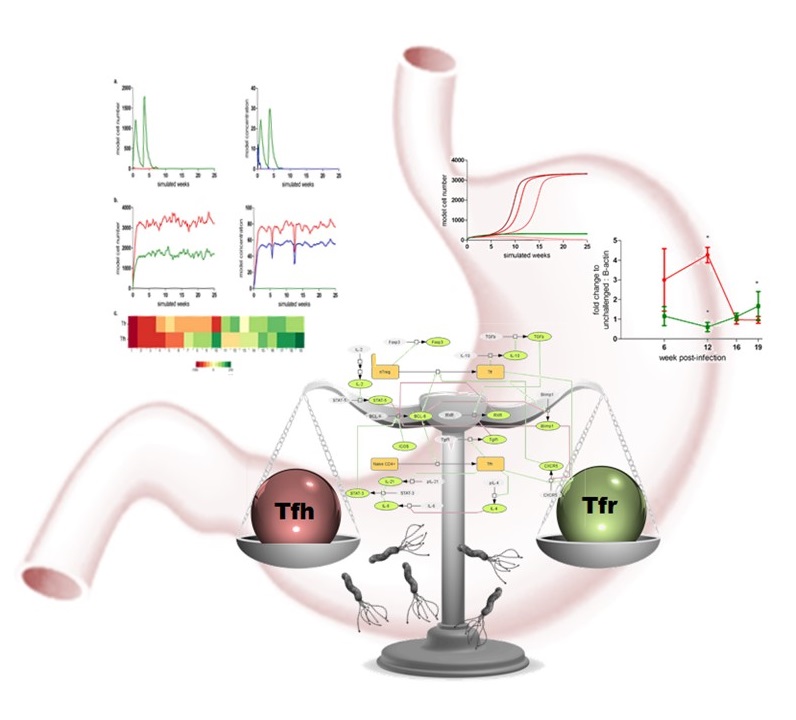
CD4+ T cells are critical controllers of the immune response capable of maintaining healthy states or promoting inflammation. In this role, certain CD4+ subsets contribute to the spontaneous inflammation in response to gastrointestinal bacteria, such as H. pylori. NIMML has recently developed a computational model that describes the network of interactions contributing to the balance between T follicular helper (TFH) and T follicular regulatory (TFR) cells. TFH and TFR produce molecules that respectively drive or prevent the expansion and accumulation of other immune cells in response to antigenic stimulation in the context of a bacterial infection.
Stability analysis and computer simulations using the new model predict that H. pylori colonization forces TFH cells into a region of bistability between regulatory and inflammatory states. In this region, the balance, while maintained, is highly susceptible to slight environment changes which would in turn cause large swings into inflammatory or anti-inflammatory tissue level outcomes leading to lesion development or maintenance of homeostasis.
“Through a modular modeling approach, in which the core network model is expanded or contracted, this work highlights the ability of a model to provide both in-depth analysis of a single scenario and the ability to explore a multitude of interactions,” said Andrew Leber, a researcher at NIMML. “We were able to answer the question: if H. pylori is present, how stable (or unstable) are these populations of immune cells. Future work will use the model to study the molecular mechanisms preventing the cells from reaching a chronically elevated state.”
At the molecular level, the model predicts that an interaction between Tgif1 and the retinoid X receptor is an important and novel controller of Tfh-Tfr differentiation. Modulation of this axis may be a therapeutic alternative to standard antibiotic treatment that removes H. pylori in cases of gastric pathology.
“This article builds on the success of our previous models of CD4+ T cell differentiation into Th1, Th2, Th17 and Treg cells. The ability of computational modeling to identify hot spots within the TFH/TFR network is a valuable resource in the identification of candidate therapeutic targets to modulate CD4+ T cell function,” said Josep Bassaganya-Riera, professor and director of NIMML at Virginia Tech. “Ongoing studies at NIMML have used ENISI MSM to combine ODE models of CD4+ T cell differentiation with agent-based tissue-level models of the immune responses to gastrointestinal pathogens bringing us one step closer to individualized precision medicine interventions for infectious and immune-mediated diseases.
For more information and results from this study, read the full article published in the Journal of Theoretical Biology.
About NIMML
The NIMML Institute is a 501 (c) (3) non-profit public charity foundation focused on a transdisciplinary, team-science approach to precision medicine at the interface of immunology, inflammation, and metabolism. The NIMML Institute team has led numerous large-scale transdisciplinary projects and is dedicated to solving important societal problems by combining the expertise of immunologists, computational biologists, toxicologists, modelers, translational researchers, and molecular biologists. The Institute is headquartered in Blacksburg, VA. For more information, please visit www.nimml.org or contact pio@nimml.org.
