Computational modeling yields a novel class of anti-inflammatory drugs
BLACKSBURG, Va., April 18, 2012 – Researchers in the Nutritional Immunology and Molecular Medicine Laboratory (NIMML) at VBI have recently discovered a new compound that shows promise in ameliorating the inflammation in the gut. Congruent to findings last year regarding abscisic acid, the discovery of Lanthionine Synthetase C-like protein 2 (LANCL2) as a novel therapeutic target could aid in relieving such diseases without the side effects of current medications. These findings have been reported in PloS ONE.
 “The long-term goal of the NIMML is to elucidate the cellular and molecular mechanisms by which novel immune therapeutics ameliorate human diseases. Our integrated computer modeling and experimental validation approaches identified LANCL2 as a novel therapeutic target for infectious, immune-mediated and inflammatory diseases,” said Dr. Josep Bassaganya-Riera, a Professor of Immunology, and the Director of the NIMML and the Center for Modeling Immunity to Enteric Pathogens (MIEP) at Virginia Tech.
“The long-term goal of the NIMML is to elucidate the cellular and molecular mechanisms by which novel immune therapeutics ameliorate human diseases. Our integrated computer modeling and experimental validation approaches identified LANCL2 as a novel therapeutic target for infectious, immune-mediated and inflammatory diseases,” said Dr. Josep Bassaganya-Riera, a Professor of Immunology, and the Director of the NIMML and the Center for Modeling Immunity to Enteric Pathogens (MIEP) at Virginia Tech.
Often the problem with current anti-inflammatory therapies is that they cause many side effects, including immunosuppression, delayed wound healing and gastrointestinal side effects such as ulcers or constipation.
“Using lessons learned from discoveries with the naturally occurring compound abscisic acid, NIMML has identified LANCL2 as a novel anti-inflammatory pathway. One compound associated with LANCL2, NSC61610, has shown great promise in suppressing gut inflammation. Our initial studies in mice indicate that the anti-inflammatory efficacy of NSC61610 depends on macrophage expression of peroxisome proliferator-activated receptor γ, a nuclear receptor downstream of the LANCL2 pathway and an internal thermostat for inflammation” said Dr. Raquel Hontecillas, an Assistant Professor of Immunology at VBI and co-director of NIMML and Immunology Lead of MIEP at Virginia Tech.
Pharmaceutical companies employ the physical large-scale, high-throughput screening of thematic compound libraries against a therapeutic target to identify new drugs. This approach is very costly and it yields mixed results.
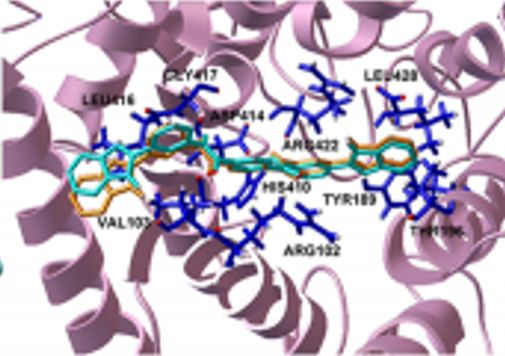
“NIMML has succeeded in the computational prediction of new ligands from large compound libraries by making use of structure-based virtual screening which is more cost-effective in drug discovery and provides reasonable hit rates. In this study, we used the structure of LANCL2 generated by homology modeling in a previous study from NIMML c. Our findings indicate that virtual screening is an effective computational-based drug design method for discovering novel anti-inflammatory drug candidates,” said Pinyi Lu, a Ph.D. student in NIMML.
This study was supported by award number 5R01AT004308 of the National Center for Complementary and Alternative Medicine at the National Institutes of Health awarded to J.B.-R., NIAID Contract No. HHSN272201000056C to JBR and funds from the Nutritional Immunology and Molecular Medicine Laboratory.
About NIMML
The NIMML Institute is a 501 (c) (3) non-profit public charity foundation focused on a transdisciplinary, team-science approach to precision medicine at the interface of immunology, inflammation, and metabolism. The NIMML Institute team has led numerous large-scale transdisciplinary projects and is dedicated to solving important societal problems by combining the expertise of immunologists, computational biologists, toxicologists, modelers, translational researchers, and molecular biologists. The Institute is headquartered in Blacksburg, VA. For more information, please visit www.nimml.org or contact pio@nimml.org.
