The NIMML Institute Announces Publication in Nature Systems Biology and Applications of a Computational Model of CD4+ T cell Immune Signaling and Metabolism to Study Novel Therapeutics
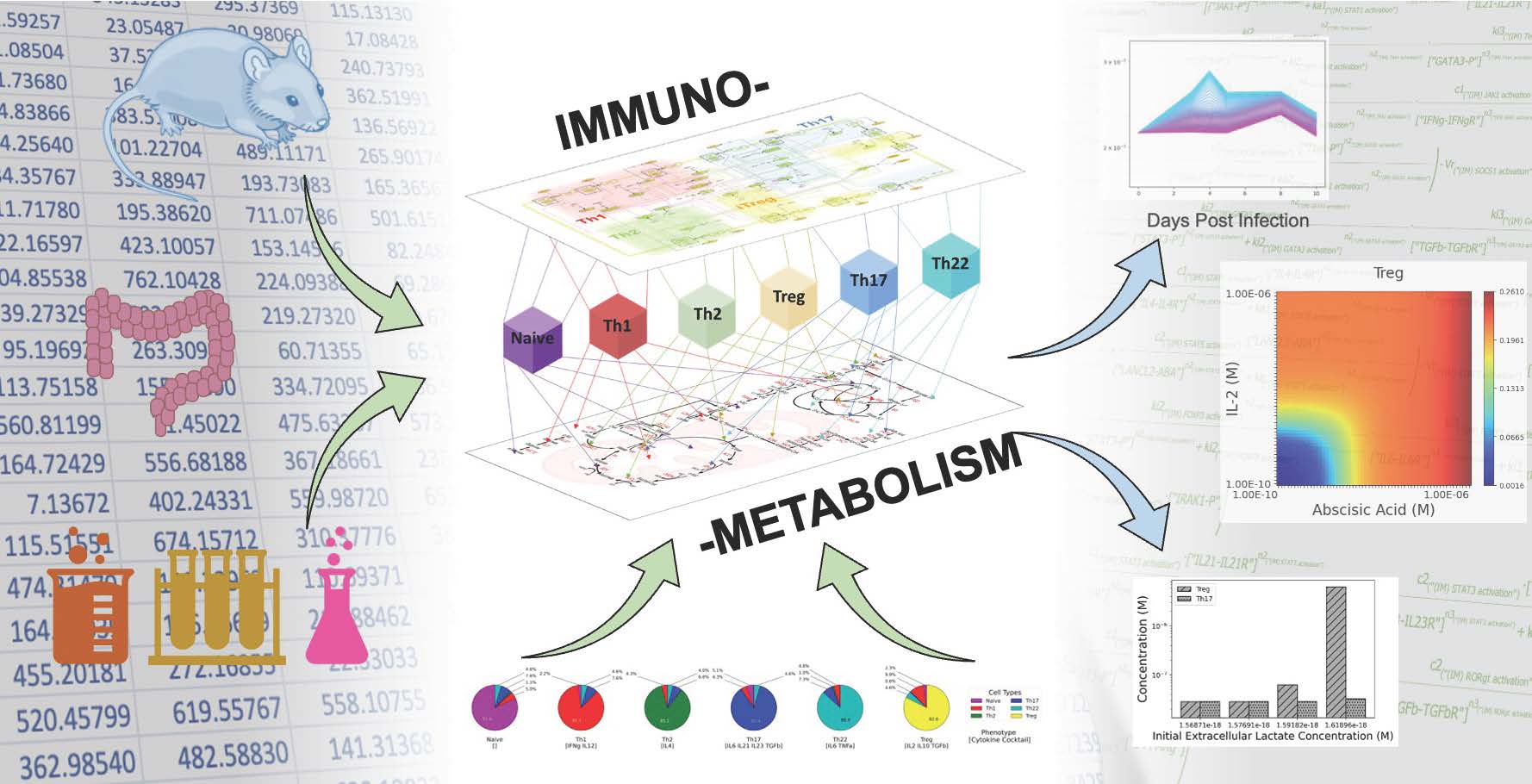
With this publication the NIMML Institute expands its library of unique computational immunology models for studying human diseases
NIMML Institute advanced computational immunology capabilities are fundamental for the launch of a new precision medicine platform anticipated for the first half of 2023.
Blacksburg, VA., November 30th, 2022 – The NIMML Institute(“NIMML”), a 501 (c) (3) non-profit foundation focused on applying transdisciplinary, team-science approaches to precision medicine, today announced the publication of a peer-reviewed article entitled “Computational modeling of complex bioenergetic mechanisms that modulate CD4+ T cell effector and regulatory functions” in Nature Systems Biology and Applications. The peer-reviewed publication applies advanced computational modeling of CD4+ T-cell function to elucidate novel immunological and metabolic mechanisms controlling how CD4+ T cells elicit effector or regulatory responses. The model can simulate clinically relevant outcomes that help identify novel drug targets for a wide range of infectious, autoimmune, metabolic, and neurodegenerative diseases. The model can also test the immunological and metabolic effects of novel therapeutics in CD4+ T cells.
When applying the computational model to the study of molecular mechanisms controlling regulatory T cell (Treg) function, our results demonstrate that activation of LANCL2 outperforms exogenous low-dose IL-2 in enhancing Treg anti-inflammatory activity. Additionally, given its narrow therapeutic range, under some conditions, IL-2 increased the inflammatory response of Th1 cells which is counterproductive in autoimmune diseases, whereas LANCL2 activation down-regulated effector responses.
“This new publication builds upon the NIMML’s track record of trailblazing discoveries under one of four nationwide Modeling Immunity for Biodefense Centers and is supported by our existing DTRA-funded R&D program to establish an advanced computational platform for rapidly developing precision immunology therapeutics at the intersection of immunity and metabolism.” said Dr. Josep Bassaganya-Riera, President and Founding Director of the NIMML Institute. “The computational models and tools developed by NIMML are disease agnostic and can be applied to the accelerated development of safe and effective therapeutics for autoimmune diseases as well as rapidly developing broad-based countermeasures against a wide range of biological, nuclear or chemical threats.”
The NIMML Institute has an extensive track record of successfully applying advanced computational immunology approaches. Several seminal papers have been published describing these results. A paper published in Gigascience details the use of agent-based modeling to identify Th17 cells as having a detrimental effect on the gut epithelium during infection by H. pylori. Other publications in Inflamm Bowel Dis. and Scientific Reports describe the development of LANCL2 as a novel therapeutic target for autoimmune disease.
The published Nature article is available at https://pubmed.ncbi.nlm.nih.gov/36418318/
About NIMML
The NIMML Institute is a 501 (c) (3) non-profit foundation focused on applying transdisciplinary, team-science approaches to precision medicine. The NIMML Institute applies its TITAN-X advanced A.I. platform to large-scale transdisciplinary projects aimed at solving important public health problems through precision medicine. NIMML combines the expertise of immunologists, computational biologists, toxicologists, computational modelers, translational and clinical researchers, and molecular biologists to translate novel scientific discoveries into medicines for human diseases. The Institute is headquartered in Blacksburg, VA. For more information, please visit www.nimml.org or contact pio@nimml.org.
