Decoding the Interface of Human Immunity and Metabolism
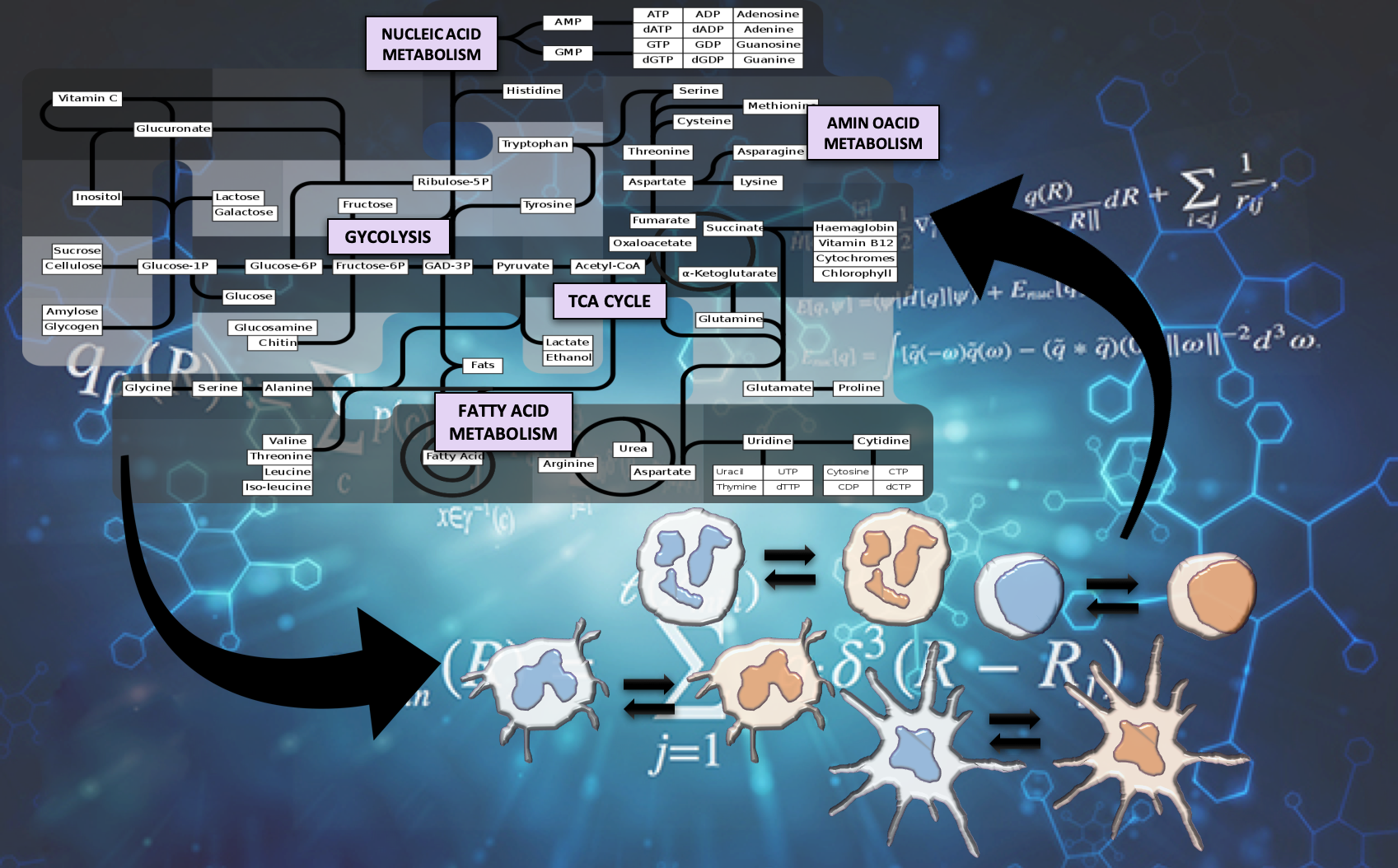
The Nutritional Immunology and Molecular Medicine Laboratory (NIMML) is pioneering the study of the immune system as a massively and dynamically interacting system plus the development of advanced computational modeling approaches to solve the complex puzzle of immunity in infectious diseases. The NIMML has built the first advanced computational model of immunometabolism.
Reprogramming immune responses through metabolic changes has been successful in immune-oncology drugs through immune checkpoint inhibitors and shows promise in the treatment of autoimmune diseases. Metabolic changes are highly linked to cell activation and effector versus regulatory functionality during immune responses to infection and vaccination. Upon initiation of the immune response to infection, activated cells undergo a significant metabolic switch to meet increased energy and structural requirements for cell proliferation. However, the immunometabolic mechanisms that control functional fates and disease outcomes during infection and vaccination remain largely unknown.
The immune system is comprised of massively and dynamically interacting cellular and molecular pathways and signals implicated in complex mechanisms that maintain immune homeostasis. A reason for pursuing computational modeling in immunometabolism is that complex networks with cooperativity and feedback exhibit nonlinear dynamics. Since nonlinear dynamical behavior is notoriously non-intuitive, researchers cannot cost-effectively predict how a given perturbation will affect system behavior over time without aid of a mathematical model. Another rationale for pursuing computational modeling in immunology and metabolism is the sheer combinatorial complexity involved in immune cell responses induced by dynamic and nonlinear cytokine, metabolic and cellular networks, and the ability to integrate information from diverse fields in a predictive manner.
The integration of the immunometabolic components, metabolites, immunological marker datasets can be used to predict distinct functional fates and clinical outcomes to compare the different treatment options and predict response to treatment. Additionally, NIMML is using Gaussian process based metamodes (stochastic kriging) to reduce the computational budget required to simulate the large complex immunometabolic models.
“Recent technological advances in computational modeling, AI and machine learning made by NIMML have given us the tools we need to embark on an ambitious endeavor of decoding human immunity. The new computational modeling of immunometabolism will help us gain new insights on metabolic reprogramming of immune cells that will pave the way for accelerating the development of safer and more effective host-based therapeutics and vaccines for emerging and re-emerging infectious diseases.” Said Josep Bassaganya-Riera, Director of NIMML.
Related press releases
Advanced Computational Modeling of the Gut for Biodefense
Immunometabolism – A Promising Gateway for Accelerated Drug Development
Applying Artificial Intelligence to Improve Drug Development
Artificial Intelligence: The Next Revolution in Healthcare and Precision Medicine
The NIMML Creates the First High-Resolution Agent-Based Model of the Gut
About NIMML
The NIMML Institute is a 501 (c) (3) non-profit public charity foundation focused on a transdisciplinary, team-science approach to precision medicine at the interface of immunology, inflammation, and metabolism. The NIMML Institute team has led numerous large-scale transdisciplinary projects and is dedicated to solving important societal problems by combining the expertise of immunologists, computational biologists, toxicologists, modelers, translational researchers, and molecular biologists. The Institute is headquartered in Blacksburg, VA. For more information, please visit www.nimml.org or contact pio@nimml.org.
